This function retrieves variant consequence types. For more details check Ensembl Variation - Calculated variant consequences.
get_variant_consequences(verbose = FALSE, warnings = TRUE)
Arguments
| verbose | Whether to be chatty about the underlying requests. |
|---|---|
| warnings | Whether to print warnings. |
Value
A tibble, each row being a variant consequence,
of four variables:
- SO_accession
Sequence Ontology accession, e.g.,
'SO:0001626'.- SO_term
Sequence Ontology term, e.g.,
'incomplete_terminal_codon_variant'.- label
Display term.
- description
Sequence Ontology description.
Details
A rule-based approach is used to predict the effects that each allele of a variant may have on each transcript. These effects are variant consequences, that are catalogued as consequence terms, defined by the Sequence Ontology.
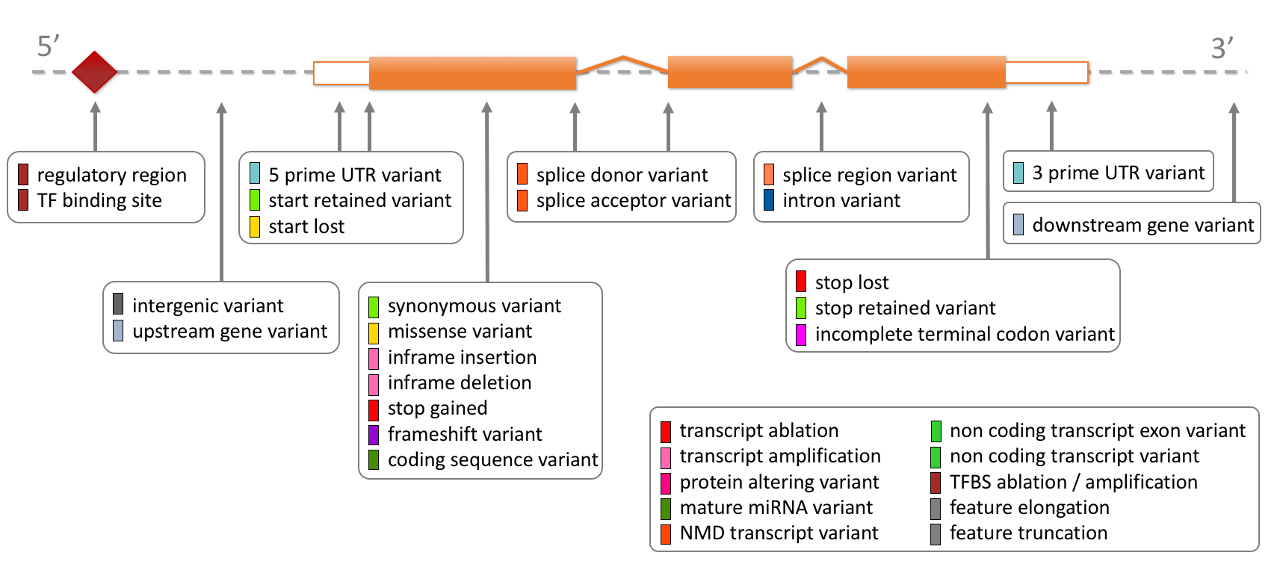
See below a diagram showing the location of each display term relative to the transcript structure:
Ensembl REST API endpoints
get_variant_consequence_types makes GET requests to
/info/variation/consequence_types.
Examples
# Retrieve variant consequence types get_variant_consequences()#> # A tibble: 37 × 4 #> SO_accession SO_term label description #> <chr> <chr> <chr> <chr> #> 1 SO:0001907 feature_elongation feature … A sequence variant … #> 2 SO:0001621 NMD_transcript_variant NMD tran… A variant in a tran… #> 3 SO:0001567 stop_retained_variant stop ret… A sequence variant … #> 4 SO:0001587 stop_gained stop gai… A sequence variant … #> 5 SO:0001626 incomplete_terminal_codon_variant incomple… A sequence variant … #> 6 SO:0001574 splice_acceptor_variant splice a… A splice variant th… #> 7 SO:0001889 transcript_amplification transcri… A feature amplifica… #> 8 SO:0001620 mature_miRNA_variant mature m… A transcript varian… #> 9 SO:0001628 intergenic_variant intergen… A sequence variant … #> 10 SO:0001627 intron_variant intron v… A transcript varian… #> # … with 27 more rows